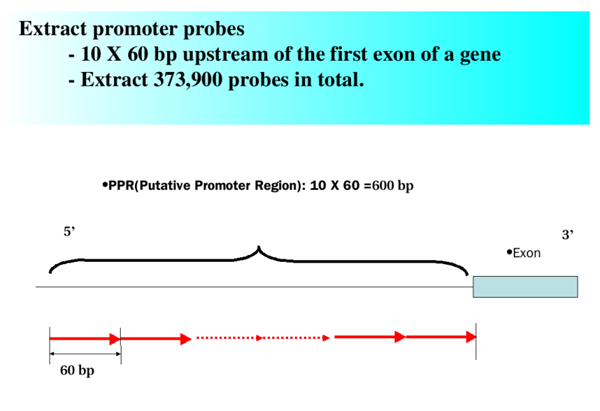
- Rice Promoter Microarray
- ChIP-CHIP experiment was conducted with Rice Promoter Microarray. The microarray was designed in conjunction with Rice Exon Microarray. The 37,400 unigenes were collected with CAP3 (http://genome.cs.mtu.edu/cap/cap3.html) by clustering 1.1 million ESTs, UniGene Build #60, down-loaded from NCBI (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene). Ten 60-nt long probes were contiguously designed in the 5' upstream region of each gene starting 600 bp ahead the start of the transcript. The microarray was manufactured at NimbleGen inc (http://www.nimblegen.com/). Random GC probes (38,000) to monitor the hybridization efficiency and four corner fiducial controls (225) were included to assist with overlaying the grid on the image.
- To assess the reproducibility of the microarray analysis, we repeated the experiment two or three times with independently prepared total RNA. The normal distribution of Cy3 intensities was tested by qqline. The data was nomalized and processed with cubic spline normalization using quantiles to adjust signal variations between chips and Rubust Multi-Chip Analysis (RMA) using a median polish algorithm implemented in NimbleScan (Workman et al., 2002. Genome Biol. 3: research0048.1 - research0048.16 ; Irizarry et al., 2003. Nucleic Acids Research 3:e15).
- Hybridization
- Genes on microarray